Optimizing Plant Defense against Pathogens
Elucidating the plant’s ability to tweak its defense response to specific pathogens.
The Science
Plants face many types of pathogens—disease-causing bacteria, viruses, and other microorganisms. In response, plants have developed sophisticated defense mechanisms. These defenses activate specific genes in response to specific pathogens. Scientists do not fully understand the mechanisms that underlie this selective activation of defense-related genes. In this new research, scientists investigated the tradeoffs in plant defenses against two pathogens. They found that Arabidopsis (thale cress) plants have a protein that regulates certain genes involved in pathogen response. This protein coordinates the responses of two plant defense pathways to create a targeted defense.
The Impact
To respond to microbial infections, plants need rapid and selective defenses. One line of defense is transcriptional reprogramming, in which specific genes are activated while others are repressed. This study identified a key genetic factor that allows plants to balance their defenses against different types of pathogens. The research also helped explain the molecular mechanism that underlies the fine-tuned response to a specific pathogen. This discovery will optimize their defense against pathogens in changing environments.
Summary
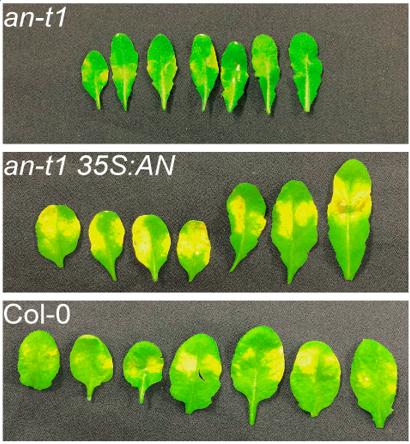
The objective of this study was to identify genetic factors and the underlying molecular mechanisms that regulate antagonism between the signaling compounds salicylic acid (SA) and jasmonic acid (JA) that result in tradeoffs in defense against pathogens. The researchers identified a plant homolog of a mammalian protein called ANGUSTIFOLIA (AN) as a key genetic factor regulating plant defense against pathogens in Arabidopsis plants. AN is a nuclear protein that regulates plant resistance towards the pathogens P. syringae and B. cinerea by specifically targeting two key genes in the pathogen response pathway. They found that co-regulation and reprogramming of gene transcription are required for AN’s regulatory role.
Contact
Jin-Gui Chen and Wellington Muchero
Biosciences Division, Oak Ridge National Laboratory
chenj@ornl.gov; mucherow@ornl.gov
Funding
This work was supported by the Office of Biological and Environmental Research within the Department of Energy (DOE) Office of Science through the BioEnergy Science Center, the Center for Bioenergy Innovation, and the Plant-Microbe Interfaces Scientific Focus Area, all at DOE’s Oak Ridge National Laboratory. The research also made use of the DOE Joint Genome Institute, a DOE Office of Science user facility at DOE’s Lawrence Berkeley National Laboratory.
Publications
Xie, M. et al. “Arabidopsis C-terminal Binding Protein ANGUSTIFOLIA modulates transcriptional co-regulation of MYB46 and WRKY33.” New Phytologist Jul 24 (2020)
Highlight Categories
Performer: DOE Laboratory , SC User Facilities , BER User Facilities , JGI