New Artificial Membranes Enable Better Understanding of Membrane Proteins
Researchers have created a novel membrane platform for studying the structure and function of membrane proteins in their realistic environment.
The Science
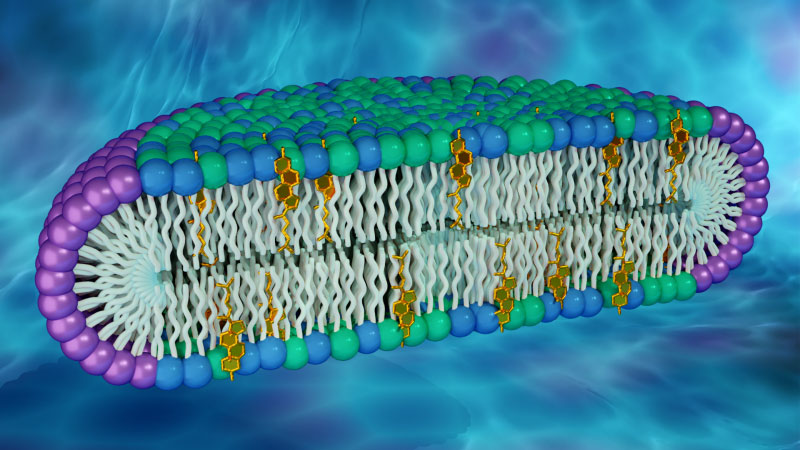
The cell membrane is the wall-like outer layer that separates the inside of a cell from its surrounding environment. The membrane mainly consists of lipids and proteins. Lipids are molecules that form the membrane’s fundamental structure. Proteins carry out important cellular functions. Scientists have now developed a disc-shaped artificial membrane that shows how proteins can exhibit different properties when embedded in membranes with different lipid compositions. The researchers used X-ray and neutron scattering techniques to confirm the synthetic membrane’s structure.
The Impact
Membrane proteins represent about one third of all human proteins, but they are hard to study. Studying the structure of membrane proteins in their realistic environment is essential to better understand their function and the role they play in diseases. The new synthetic membrane provides a novel platform to appropriately vary the types of lipids in a membrane. This ability helps researchers to better understand different membrane proteins in a realistic environment.
Summary
Several kinds of lipid molecules are found in cell membranes, and certain lipids have the tendency to aggregate into clusters, known as rafts. Researchers believe that proteins could behave differently in lipid raft environments, compared to non-raft regions in a membrane, but this hypothesis has not been fully evaluated. One reason is that membrane models used to study membrane proteins rarely contain rafts. This deficiency prompted a team of scientists to develop a new membrane model that is abundant in two types of lipids known to form rafts in cell membranes: namely, cholesterol and sphingomyelin. Using the artificial membrane, the team found notable differences in the structure and dynamics of a protein when exposed to a raft forming lipid composition compared to a non-raft forming membrane.
The researchers characterized the new membrane model using a combination of small-angle X-ray scattering (SAXS) and small-angle neutron scattering (SANS). Access to the Bio-SANS instrument at the High Flux Isotope Reactor (HFIR) and the Bio-SAXS instrument (LiX) at the National Synchrotron Light Source II (NSLS II) was provided by the facilities’ SAXS/SANS Joint Access Program. Using both methods, the scientists were able to generate a more accurate picture of the membrane’s morphology.
Contact
John Katsaras
Oak Ridge National Laboratory
katsarasj@ornl.gov
Charles Sanders
Vanderbilt School of Medicine Basic Sciences
chuck.sanders@vanderbilt.edu
Funding
This work was supported by the National Institutes of Health, the National Science Foundation, Vanderbilt University, European Union Horizon 2020, Deutsche Forschungsgemeinsachft, and The Department of Energy Office of Science, Office of Biological and Environmental Research and Basic Energy Sciences (for support of the beamlines and operation of the HFIR and NSLS II, DOE Office of Science user facilities).
Publications
J.M. Hutchison, et al. “Bicelles Rich in both Sphingolipids and Cholesterol and Their Use in Studies of Membrane Proteins.” Journal of the American Chemical Society, 142, 29, 12715–12729 (2020). [DOI: 10.1021/jacs.0c04669]
Related Links
Novel cell membrane model capable of uncovering new protein properties, Oak Ridge National Laboratory, Neutron Sciences
Highlight Categories
Performer: University , DOE Laboratory , SC User Facilities , BES User Facilities , NSLS-II , HFIR
Additional: Collaborations , Non-DOE Interagency Collaboration