Watching the Evolution of a Protein’s Function
Study reveals structural changes leading to catalytic activity.
The Science
A major challenge in research to enable large-scale biofuels production is developing enzymes that are highly efficient in converting biomass components into usable fuels.
The Impact
Using directed evolution (i.e., a technique for modifying protein function), researchers have determined the structural basis for converting a noncatalytic small protein into an effective enzyme for linking RNA molecules.
Summary
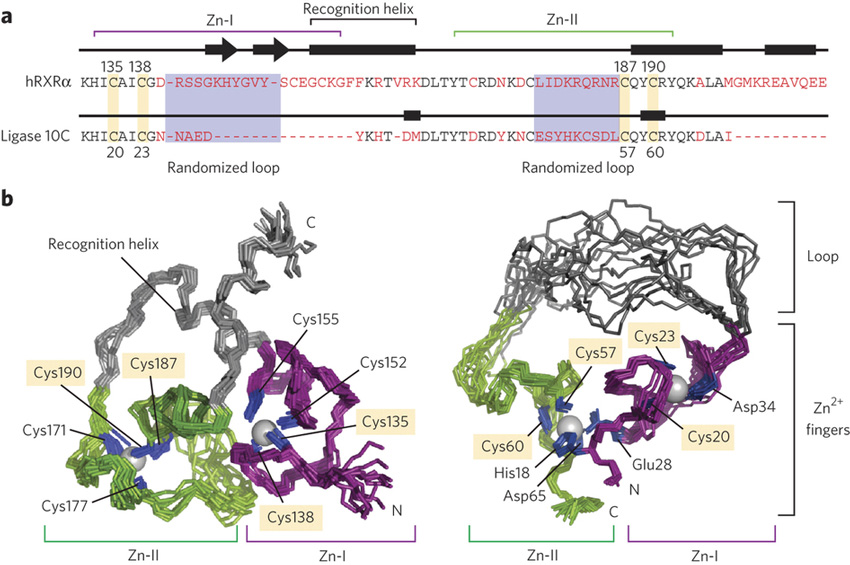
Enzymes are proteins structurally configured to catalyze conversions of substrates to products. Thousands of protein structures are known, including those of many valuable enzymes. However, much less is known about how small changes in a protein’s composition can alter its three-dimensional structure, control its catalytic efficiency, or even convert a protein with no catalytic function into an efficient catalyst. Scientists in this study used an extended X-ray absorption fine structure (EXAFS) station at the Stanford Synchrotron Radiation Lightsource to determine the active-site structure of a newly synthesized enzyme. The EXAFS experiments were able to show the exact chemical environment of each zinc atom in the new enzyme, leading to an explanation of why it developed the catalytic activity.
Contact
Burckhard Seelig
Department of Biochemistry, Molecular Biology and Biophysics, University of Minnesota, Minneapolis Technology Institute, University of Minnesota, St. Paul
seelig@umn.edu
Funding
This work was supported by the U.S. National Aeronautics and Space Administration (NASA) Agreement no. NNX09AH70A through the NASA Astrobiology Institute–Ames Research Center, the Minnesota Medical Foundation, and the U.S. National Institutes of Health (NIH; T32 GM08347, T32 DE007288 GM100310, and P41 RR001209). Stanford Synchrotron Radiation Lightsource (SSRL) operations are funded by the U.S. Department of Energy (DOE) Office of Basic Energy Sciences. The SSRL Structural Molecular Biology program is supported by the NIH National Center for Research Resources and the DOE Office of Biological and Environmental Research.
Publications
Chao, F.A., et al. “Structure and dynamics of a primordial catalytic fold generated by in vitro evolution.” Nat. Chem. Biol. 9 (2), 81–83 (2013). [DOI: 10.1038/nchembio.1138].
Highlight Categories
Program: BER , BSSD , BES , SUF
Performer: University , DOE Laboratory , SC User Facilities , BES User Facilities , SSRL